Welly to the wescue
/I apologize for the widiculous title.
Last week I described some headaches I was having with well data, and I introduced welly, an open source Python tool that we've built to help cure the migraine. The first versions of welly were built — along with the first versions of striplog — for the Nova Scotia Department of Energy, to help with their various data wrangling efforts.
Aside — all software projects funded by government should in principle be open source.
Today we're using welly to get data out of LAS files and into so-called feature vectors for a machine learning project we're doing for Canstrat (kudos to Canstrat for their support for open source software!). In our case, the features are wireline log measurements. The workflow looks something like this:
- Read LAS files into a welly 'project', which contains all the wells. This bit depends on lasio.
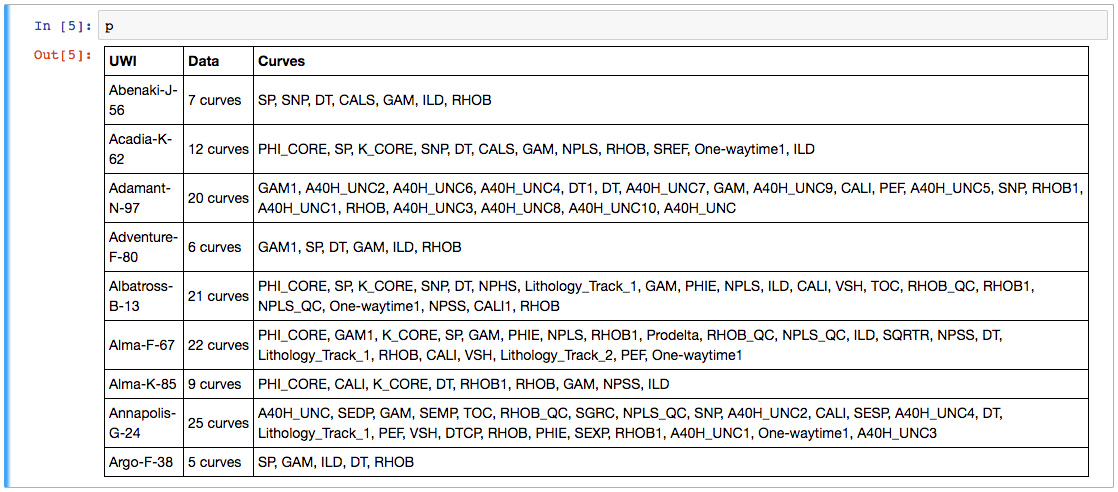
- Check what curves we have with the project table I showed you on Thursday.
- Check curve quality by passing a test suite to the project, and making a quality table (see below).
- Fix problems with curves with whatever tricks you like. I'm not sure how to automate this.
- Export as the X matrix, all ready for the machine learning task.
Let's look at these key steps as Python code.
1. Read LAS files
from welly import Project
p = Project.from_las('data/*.las')2. Check what curves we have
Now we have a project full of wells and can easily make the table we saw last week. This time we'll use aliases to simplify things a bit — this trick allows us to refer to all GR curves as 'Gamma', so for a given well, welly will take the first curve it finds in the list of alternatives we give it. We'll also pass a list of the curves (called keys here) we are interested in:
The project table. The name of the curve selected for each alias is selected. The mean and units of each curve are shown as a quick QC. A couple of those RHOB curves definitely look dodgy, and they turned out to be DRHO correction curves.
3. Check curve quality
Now we have to define a suite of tests. Lists of test to run on each curve are held in a Python data structure called a dictionary. As well as tests for specific curves, there are two special test lists: Each and All, which are run on each curve encountered, and on all curves together, respectively. (The latter is required to, for example, compare the curves to each other to look for duplicates). The welly module quality contains some predefined tests, but you can also define your own test functions — these functions take a curve as input, and return either True (for a test pass) for False.
import welly.quality as qty
tests = {
'All': [qty.no_similarities],
'Each': [qty.no_monotonic],
'Gamma': [
qty.all_positive,
qty.mean_between(10, 100),
],
'Density': [qty.mean_between(1000,3000)],
'Sonic': [qty.mean_between(180, 400)],
}
html = p.curve_table_html(keys=keys, alias=alias, tests=tests)
HTML(html)the green dot means that all tests passed for that curve. Orange means some tests failed. If all tests fail, the dot is red. The quality score shows a normalized score for all the tests on that well. In this case, RHOB and DT are failing the 'mean_between' test because they have Imperial units.
4. Fix problems
Now we can fix any problems. This part is not yet automated, so it's a fairly hands-on process. Here's a very high-level example of how I fix one issue, just as an example:
def fix_negs(c):
c[c < 0] = np.nan
return c
# Glossing over some details, we give a mnemonic, a test
# to apply, and the function to apply if the test fails.
fix_curve_if_bad('GAM', qty.all_positive, fix_negs)What I like about this workflow is that the code itself is the documentation. Everything is fully reproducible: load the data, apply some tests, fix some problems, and export or process the data. There's no need for intermediate files called things like DT_MATT_EDIT or RHOB_DESPIKE_FINAL_DELETEME. The workflow is completely self-contained.
5. Export
The data can now be exported as a matrix, specifying a depth step that all data will be interpolated to:
X, _ = p.data_as_matrix(X_keys=keys, step=0.1, alias=alias)That's it. We end up with a 2D array of log values that will go straight into, say, scikit-learn*. I've omitted here the process of loading the Canstrat data and exporting that, because it's a bit more involved. I will try to look at that part in a future post. For now, I hope this is useful to someone. If you'd like to collaborate on this project in the future — you know where to find us.
* For more on scikit-learn, don't miss Brendon Hall's tutorial in October's Leading Edge.
I'm happy to let you know that agilegeoscience.com and agilelibre.com are now served over HTTPS — so connections are private and secure by default. This is just a matter of principle for the Web, and we go to great pains to ensure our web apps modelr.io and pickthis.io are served over HTTPS. Find out more about SSL from DigiCert, the provider of Squarespace's (and Agile's) certs, which are implemented with the help of the non-profit Let's Encrypt, who we use and support with dollars.













 Except where noted, this content is licensed
Except where noted, this content is licensed